Archive for Module1
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
Contents
The ENA database
The European Bioinformatics Institute (EBI) hosts the ENA (European Nucleotide Archive) database: one part of ENA is called EMBL-bank, containing annotated primary sequence data. The other two parts are the Trace Archive and the Short Read Archive (SRA), containing batch-submitted primary sequence data.
EBI has multiple search portals:
Note: information is liquid. Records change all the time: info is removed and added. Therefore, screenshots may not be up-to-date.
The ENA Browser
Go to the ENA Browser. You see two text field, the upper one for "Text search", the lower for "Sequence search". We will concentrate on the text search, sequence searches will be covered in Module 2. You can search using free text (e.g. species names, disease names, feature names,...) or using an accession number.
Exercise 1: caspase
Perform a search for 'caspase complete cds'.
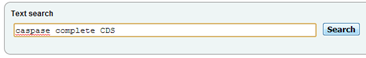
The ENA search returns records from the EMBL-bank part of ENA, divided into "Update" and "Release". "Update" contains records that were recently updated. Clicking the "+" sign expands the corresponding section, revealing the individual search results.
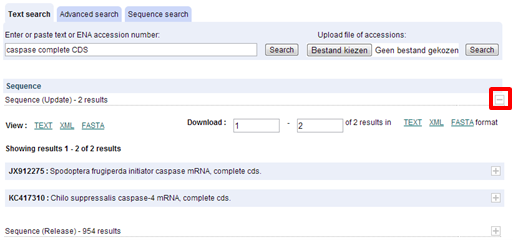
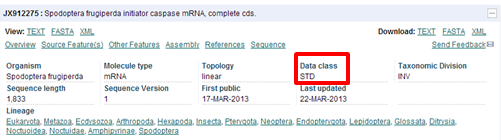
Each record can be further expanded by clicking on the "+" sign to see more details of the record. Do this for the first record of the "Update" list (JX912275 : Spodoptera frugiperda initiator caspase mRNA, complete cds).
| Which data class does this ENA record belong to? |
|---|
| The record belongs to the STD class. |
The most useful entries with the most relevant annotations are from the 'STD' (standard) data class. See more info on ENA database structure.
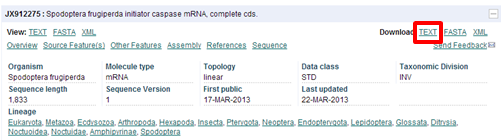
| Download the record in FASTA format? |
|---|
| In the "Download" section click on "FASTA" This will create a file called "ena.fasta" in the "Downloads" folder of your computer. Open the file in WordPad. |
| Can you tell the major difference between a sequence stored in FASTA and a sequence in EMBL text format? |
|---|
| FASTA has been stripped of all annotations: it is basically just the sequence, and one description line (corresponds to the 'DE' line in the EMBL text file). |
Exercise 2: kinase
The nicest thing about ENA Browser search, is the fact that the results are categorized by the part of ENA from which they originate. This becomes clear when you do a text search with "kinase".
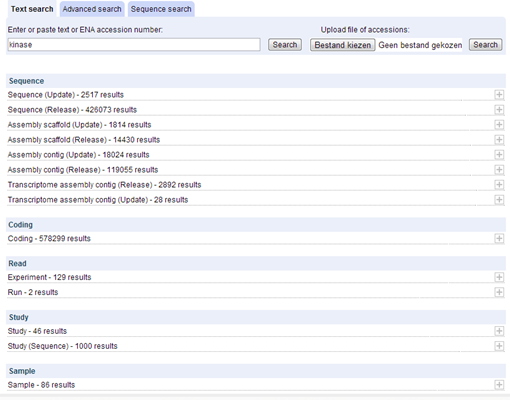
The results page groups the entries according to type of sequence.
The text searches that you can perform using the ENA Browser are very 'crude'. For example, when you search for "kinase", every record containing somewhere the word "kinase" is shown, even non-kinase sequences just as in Genbank. Be aware of this because this is often not what you want!
EBI Search, cross-database search at EBI
EB-eye is a cross-database search tool for EBI databases similar to Entrez for NCBI databases. You can access it on EBI Search.
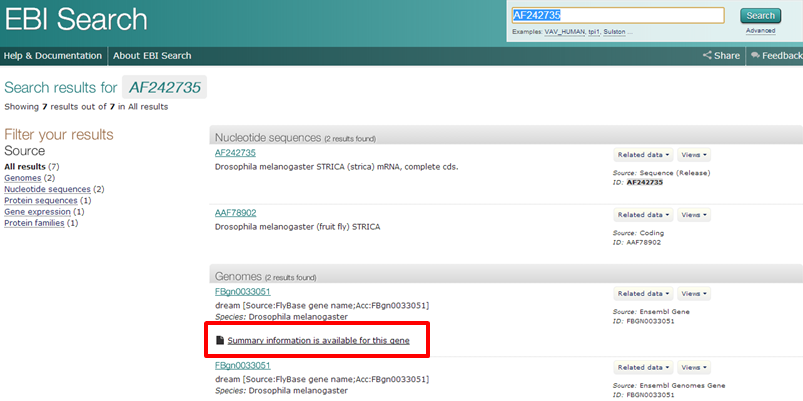
Exercise 1: AF24735
This redirects you to the EBI summary record of this gene
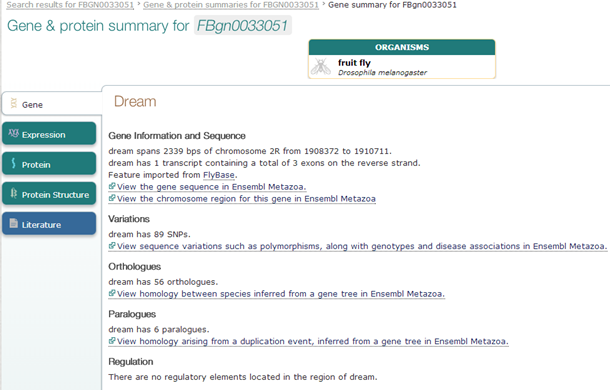
EBI provides very nice overview pages, with links to many other databases. A good place to start.
MRS
NCBI, ENA end UniProt have all their own search engines. Would it not be convenient to have one search portal searching all databases?
MRS (Maarten's Retrieval System) is a such webservice! Using MRS, you can access directly many different sequence databases at once. As a bonus, MRS handles synonyms and misspelled words very good by offering alike words for your search term.
MRS: performing a simple search
Go to http://mrs.cmbi.ru.nl. The search box at the top provides you access to different databanks with different content and from different origin (more info on which databases are available in MRS). MRS is ideally suited to do advanced searches, but let's start with simple. :-)
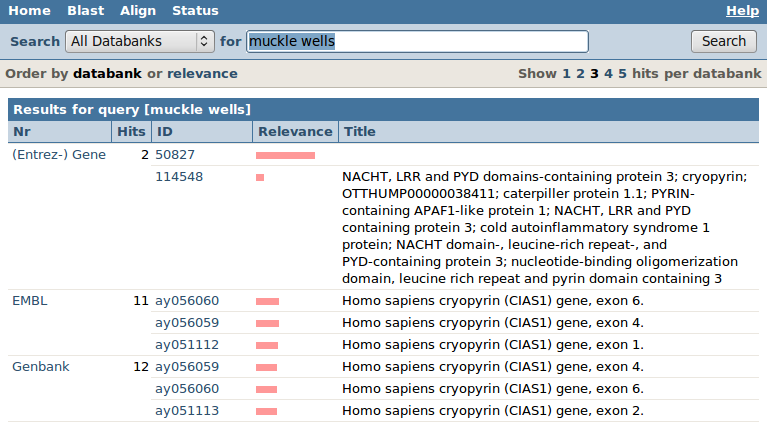
Let's see if we can find some information about the Muckle-Wells syndrome, a human genetic disease. In the for box type "(muckle-wells) OR (muckle wells)" and click on the Search button on the right.
You will get a list of databanks, with for each databank how many "hits" were found, as well as some (!) of the hits (by default 2). You should see a series of sequence databanks as well as some other non-sequence databanks like OMIM or Entrez Gene.
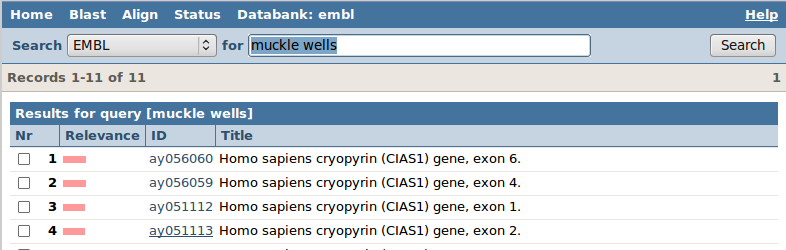
Let's first look at the hits found in the sequence database EMBL: click on EMBL. You will get the EMBL entries matching your query. The ID column is the accession number of that sequence, the title gives a description.
The pink bar measures the "relevance" of the hit (based on how often your query words "muckle" and "wells" occur in the text of the entry versus in the complete EMBL databank). Click on the accession number "af410477". General browsing tip: press ctrl+F to open a search box in your browser, and enter the search term af410477.
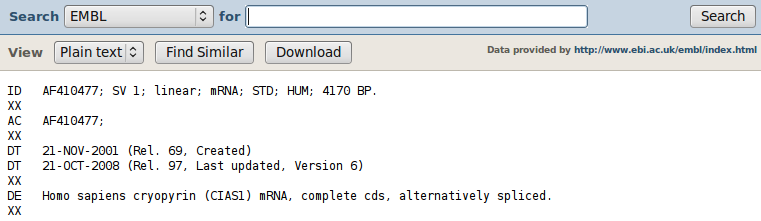
Now we see all information from entry AF410477. MRS has displayed it in a "nice" way, but you can also have a look at the "raw" text: set the selector "View" to "Plain text" (in this case, EMBL format). Recapitulation: each line start with an index, the FT part is the largest and contains annotation information and cross-references to other databases. Using these indexes, relevant information is structured and hence can be easily searched by humans and computers.
For now, go back to the nice version ![]() .
.
We have already introduced cross-references ('db_xref'): in the "nice" version most of them are displayed as hyperlinks, taking you directly to the linked database. For example, look for the coding sequence of this entry "af410477". What is a coding sequence? In bioinformatics, it is an annotation to a sequence, right? Hence you can find this under features (FT) section, more specifically the feature called CDS. For the CDS feature, some cross-references are present, after the qualifier db_xref.
| In record AF410477, one of the features is called 'source'. It shows info about where this sequence originates from. Check the important and only qualifier db_xref. Here is referred to an important database: which one? |
|---|
| Search for the record with this accession number in MRS. |
| Under FT, source details of this sequence are available. Click on db_xref taxon:9606. |
| A record from NCBI's taxonomy database is shown. Every species and branch in the phylogenetic tree of life has a taxon id, human being id 9606. All sequences are linked to a taxon id from NCBI's database, referring to a certain species. You can search on this term! |
MRS: performing a more refined search
The advanced search depends on indexes of the entries. Not all info of entries is indexed: which info is indexed, can be seen from the database summary page. This indexing is a commonly used technique to speed up database searches and to fine tune your searching. Let's give an example.
I want you to get all the proteins of the Porcine Adenovirus. Let's do naive! Type in 'Porcine Adenovirus' in MRS, using UniProtKB. You see that it will not dproduce right results! You cannot rely on these results, they give you too much redundancy.
| Think about why this is. |
|---|
| Indeed, every record that has somewhere porcine and adenovirus in their annotation, is reported... Not what we want! |
So we need a better approach. Let's first set up our approach. What are all the proteins of the Porcine Adenovirus? These are all the gene product of the genes on the genome of Porcine adenovirus. So can we make use of the annotated genome sequence genome? Yes! From the genome annotations, we can retrieve the cds annotations to finally obtain all proteins. What we do:
- we need the accession number of the genome.
- Next we exploit the fact that databanks contain cross-references to each other, so that we can search for all UniProt entries (UniProt = the major protein database), which reference the EMBL entry from step 1 containing the genome. As such, we retrieve all protein sequences from porcine adenovirus.
Step 1:
| - We search for the complete genome of porcine adenovirus, with making use of the appropriate indices. |
|---|
| A genome sequence is a DNA sequence: hence we look for an EMBL record. Select with the Search selector "EMBL". |
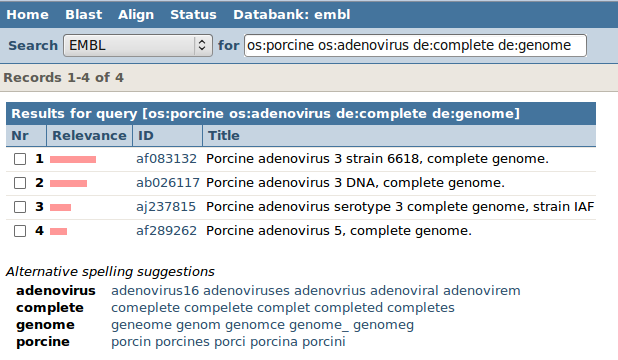
| The index for organism is OS (as for 'organism source'). Let MRS know that organism need to be porcine adenovirus, but typing "os:porcine os:adenovirus". To retrieve the complete genome, we search in the description (index "DE") for the complete genome, by typing: de:complete de:genome". |
| How do we know the "OS" and the "DE"?? You can look up in MRS which fields are indexed per database. Also information is displayed on a line can be found there. Use it to your advantage: check the os and de field for EMBL! |
| Search! You will probably get 4 entries, corresponding to 4 different strains. |
MRS combines search terms with AND by default. You can explicitly use AND OR NOT, or use the symbols & | !
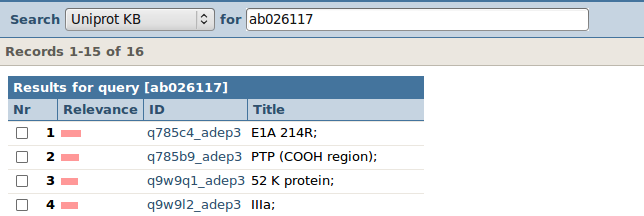
Let's continue with ab026117. You can convince yourself that the words "porcine" and "adenovirus" and "complete" and "genome" can indeed be found in the "OS" and "DE" tagged fields. This genome contains coding sequences (CDS) for 16 proteins. Remember, we only needed the accession number of the genome: so take 'ab026117' with you to the next step.
Step 2
| - Fetch all protein sequences which all cross-referenced to the porcine adenovirus genome. |
|---|
| Nice trick! Simply search for the accession number of the genome (ab026117) in the Uniprot KB database. The search will find these back in the db_xref fields. |
| We get a list of the 16 proteins encoded by this genome. |
Perhaps you can solve the next question, using the same techniques, by yourself. Remember: decide which database to use, which sequence you want, which indexes you need.
| Retrieve the complete cds from falcipain, a cysteine protease from Plasmodium falciparum. Combine different search tags for this. |
|---|
| The complete CDS is a nucleotide sequence: we can search EMBL for this. Note: remember also Unigene, the db containing all transcripts from one gene. Unfortunately, Unigene currently has no records for Plasmodium falciparum, as no hits will return after searching for it in Unigene. |
| 'Complete CDS' is always mentioned in the descriptor field. The organism field we know alreadyfrom previous exercise. If not, you can look it up here. |
| Following query should do the trick: os:'Plasmodium' os:'falciparum' de:'complete' de:'cds' falcipain. |
| Aha! 11 entries are displayed. Click on EMBL to see all hits. Redundancy! Now it is up to you which entries you are interested in. |
For your gene of interest ...
For your gene of interest, can you:
- find the refseq nucleotide sequence?
- find all primary sequence information back?
- if it is protein coding, retrieve the 50% cluster of protein sequences at Uniprot?
Conclusions of these exercises
This wraps up our introduction to searching and downloading sequences from sequence databases.
- Information is stored in a structured way in databases,
- which allows rapid searching in databases.
- Knowing the structure is needed for an optimal search strategy.
MRS gives you the power to query from a single access point many different databases. Carefully constructing and typing the query can be cumbersome: therefore, graphical interfaces, such as Entrez or ENA search provide a user-friendly portal for searching.
Optional exercises
Extracting sequence parts corresponding to annotations
Annotations are very useful information. You might sometimes want to extract sequences corresponding to annotations. This can for example be done with SMS2, the Sequence Manipulation Suite , which is accessible through our website: BITS software --> Hosted Apps --> SMS2 (mirror), or directly by clicking here. As an example, sequence with accession number NM_010111 contains a lot of annotations, of which we are interested in the Sequence Tagged Sites (STS). We want to extract them in fastA format.
- Download NM_010111 in Genbank format
- Use SMS2 --> GenBank Feature Extractor to retrieve all STS sequences (paste the sequence in the box and click on submit).
- Copy/paste the sequences starting with STS (use search function of your browser).
Changing sequence file formats
We have seen many sequence file formats, in which metadata is documented in different ways. Not all tools accept all formats, causing a lot of trouble sometimes. SMS2 can convert you sequences between fasta, genbank and embl.
Overview of the most important databases in MRS
Since MRS is a metasearch engine of different databases, it has this nice feature of MRS: if you want more information about database currently accessible by MRS, you can click the button on top of the page, showing Databank: uniprot or ![]() ,... Or you can go to the complete overview by clicking on
,... Or you can go to the complete overview by clicking on ![]() on the top bar.
on the top bar.
| How many entries are available in Swiss-Prot? |
|---|
| By checking the database statistics in MRS, 'sprot' has 532,792 entries (on 4 Nov 2011) |
An example of a specialized sequence databank : search for an old vector sequence that you cannot easily find elsewhere
The sequence of the expression vector pMAL-c has never been submitted to
EMBL/GenBank/DDBJ. You can however find it in the Intelligenetics Vector databank, which has been composed from various sources. It has not been updated
anymore since 1996 but is still interesting for finding some old vector sequences you can find nowhere else. The databank can be downloaded from the NCBI anonymous
ftp server. There are also a few sites where you can search it on-line : the MRS server of the Belgian EMBnet Node (which will however soon disappear), the SRS server of the DFKZ (German Cancer Research Center) and the Web site of the University of Stanford.
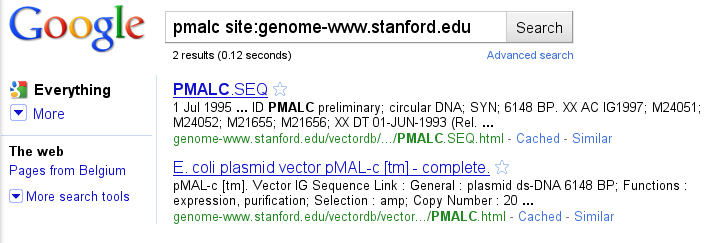
Go to http://genome-www.stanford.edu/vectordb and follow the links "Plasmid Vectors" and "Complete Sequences Starting PL-PS". You will however note that the vector we search is not in the page, presumably because of human error. We can however reach our goal by an unexpected route, exploiting the fact that the Google Web crawler reads and indexes the pages on the U. Stanford Web site! Go to http://www.google.be follow the link "Advanced search", type pmalc in the "all these words" box, type genome-www.stanford.edu in the "Search within a site or domain" box and click on "Advanced Search".
Hop, you should have found an entry with entry name PMALC and accession number IG1997. Note that there are some modern vectors that you can find nowhere except maybe inthe release notes of the company that makes them.
Taxonomies
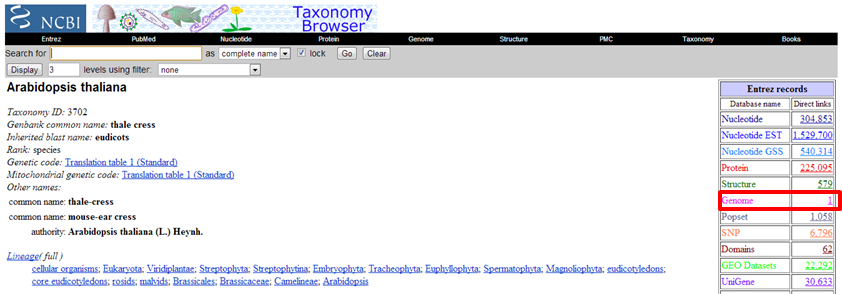
Taxonomy is an NCBI database. It is the one and only reference for species classification of all sequences. You can search this database via the Entrez portal http://www.ncbi.nlm.nih.gov/Entrez. Search for "hiv" and click through until you get to a Taxonomy record. Every species and node on the taxonomical lineage receives a Taxonomy ID. In bioinformatics terms, a species is referred to by this ID. From the Taxonomy database, you can retrieve sequences per species, which is sometimes very convenient.
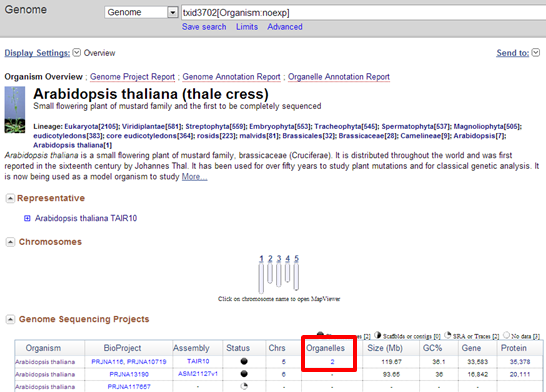
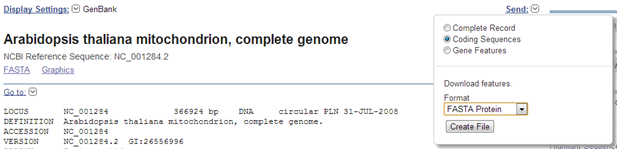
Exercise 1: Arabidopsis mitochondrial genome
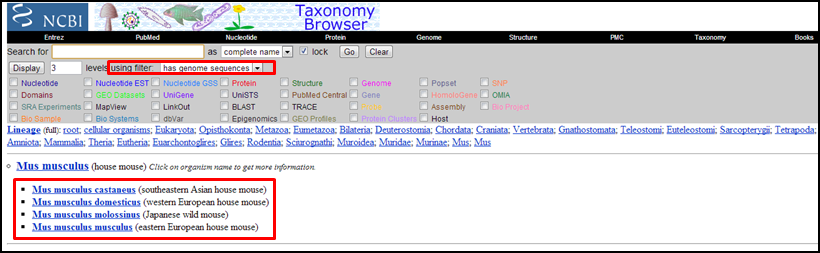
Exercise 2: mouse genome sequences
Find taxonomy info for mouse using Entrez.
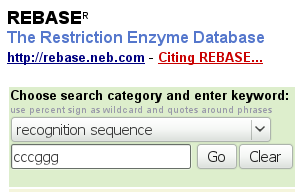
The Rebase databank
Rebase is a databank with information about restriction enzymes, maintained by R. Roberts at New England Biolabs, a company that sells, among other things, restriction enzymes. Suppose you need for one of your experiments an enzyme specific for the sequence CCCGGG. Go to http://rebase.neb.com/rebase/. Make sure the "choose search category..." selector stands on "recognition sequence", then type cccggg in the box underneath and click "Go".
You should obtain a list with more than 20 restriction enzymes, as well as some methyltransferases. Look at one of the restriction enzymes. Note the "Prototype" field and follow the hyperlink to SmaI. Of all enzymes specific for CCCGGG, SmaI is the most readily available or otherwise the "most representative". Note that SmaI makes blunt ends ; some other enzymes in the list cut in a different position or the exact location of the cut is not known. If you click on "Similar enzymes" you get a complete list.
You can obtain a list of the companies where you can order SmaI by following the "Commercially Available" link in the SmaI page or the link below "Suppliers" in the Similar Restriction Enzymes page.
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training