Ex 1 - Basic operations in IPA
Go to parent Ingenuity Pathway Analysis training
Contents
Starting IPA and searching
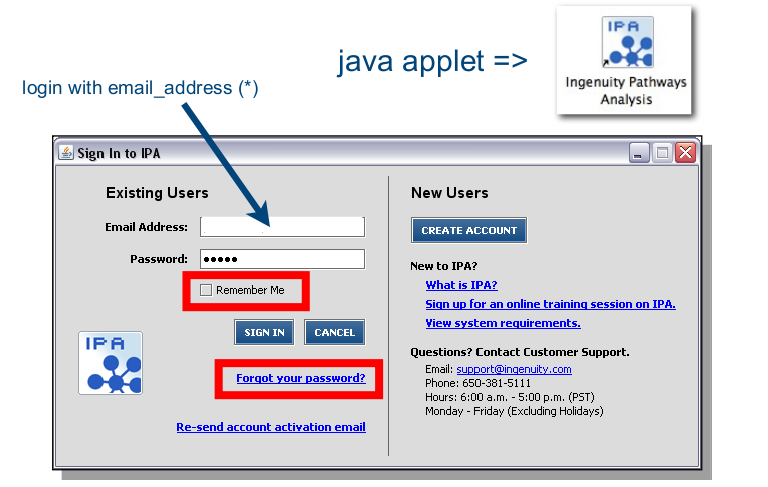
Go to https://www.ingenuity.com/pa and login.
* Back in your lab, ask your BITS commitee member for login credentials
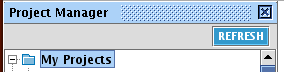
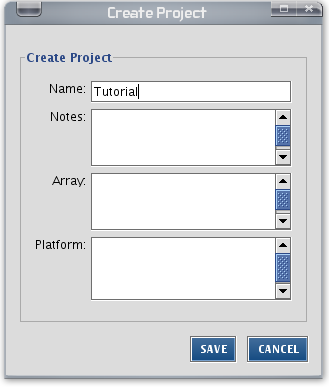
When IPA is started, make a new project folder in IPA for this training session. You can do this by going to File -> New -> Project... (or right-click in the project manager and choose New Project. Name your new folder 'Tutorial' or any name that makes sense.
From now on, save everything in your Tutorial project folder.
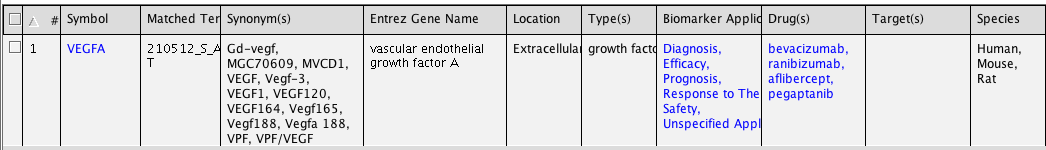
Search for an affymetrix probe (210512_s_at), a gene symbol (VEGFA), a genbank accession (NM_001025366) ...
Use the simple SEARCH for this.
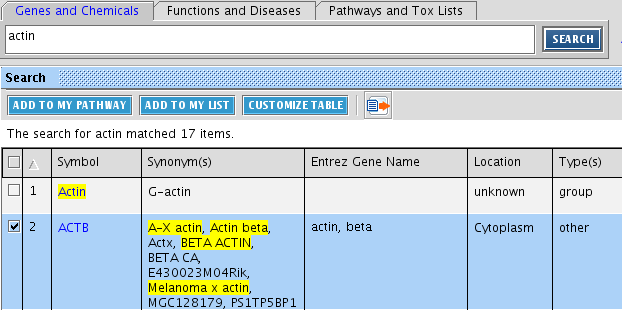
Note the three tabs above the search box. You can search for genes or chemicals, functions or diseases, and Canonical pathways. Any of these searches will result in a list of molecules. Type in in 'Genes & Chemicals' the search term. You should see following output, with information about the entity summarized. Clicking on VEGFA leads to a webpage with Ingenuity information about this entity. You can study this information. Repeat this with your gene of interest
Tip: the search box also accepts more than one entity name
IPA accepts many identifiers and is therefore suited to convert one list of identifiers to another.
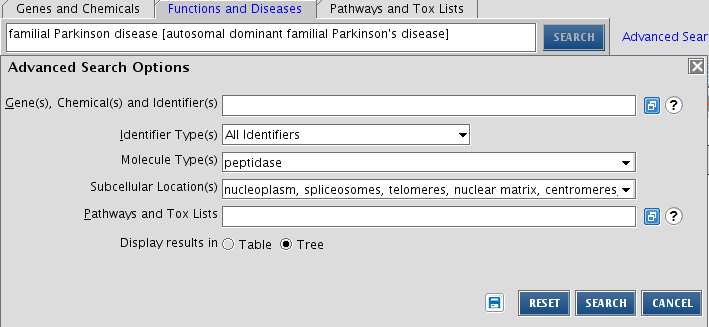
Search for peptidases found in the nucleus and implicated in Parkinson’s disease
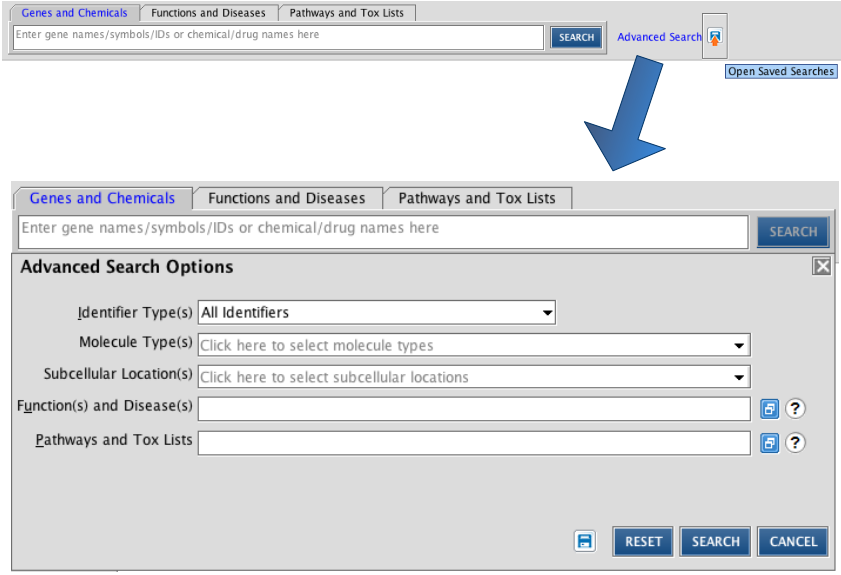
For a more detailed search, click the advanced search button. Several parameters can be given here to restrict your search. These parameters are based on the ontology of IPA.
To answer the question, make use of Advanced Search, go to the tab Functions and Diseases (as Parkinson's disease is the main focus). In the search field, start typing Parkinson disease and pick the right description from the autocompleted list that you observe. Fill the other details in as shown below. Note that you have to pick the specifications from a list. The results can be displayed in table format (as is used in the simple search), or in a tree format.
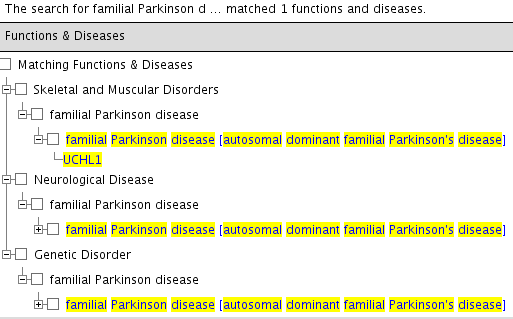
Which should give following output (in tree format):
Your first pathway
A Pathway in IPA is a network consisting of molecular entities (Genes or Chemicals - the lowest level in the IPA scheme) which are related to one another through different relationships. Each entity has its own symbol derived from its function (see the Quick Ref sheet from BITS). Entities directly connected to each other are called 'neighbors'.
QUESTION Get the full neighbourhood of your gene of interest
First search your gene you are interested in by typing its name in the 'Genes and Chemicals' search box. From the resulting list, select your gene by ticking it. We demonstrate this with the ACTB gene. You can use any gene you want.
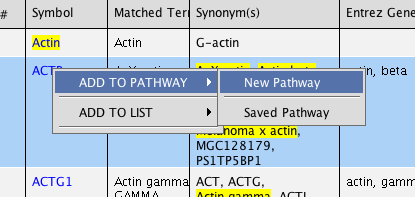
Click on ADD TO MY PATHWAY button, or right-click and select ADD TO PATHWAY > New Pathway.
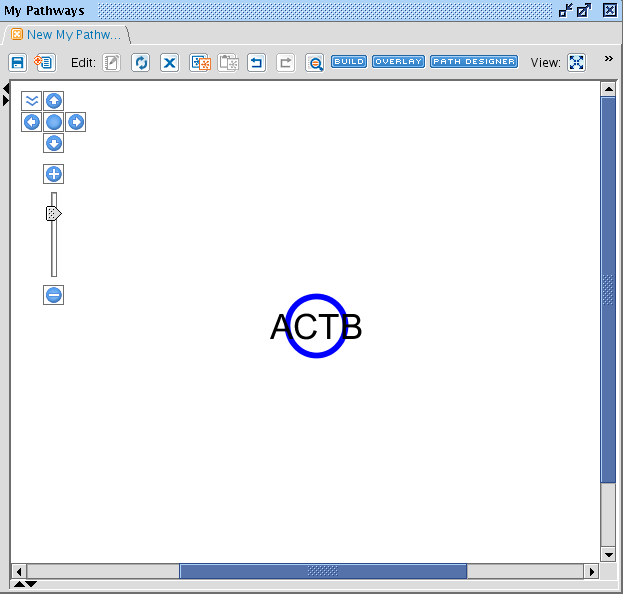
When you have done this, the Pathway view is opened and your molecule is displayed.
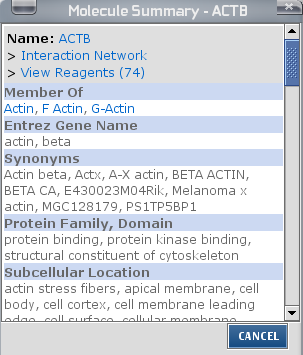
By double-clicking on the entity, more information is displayed.
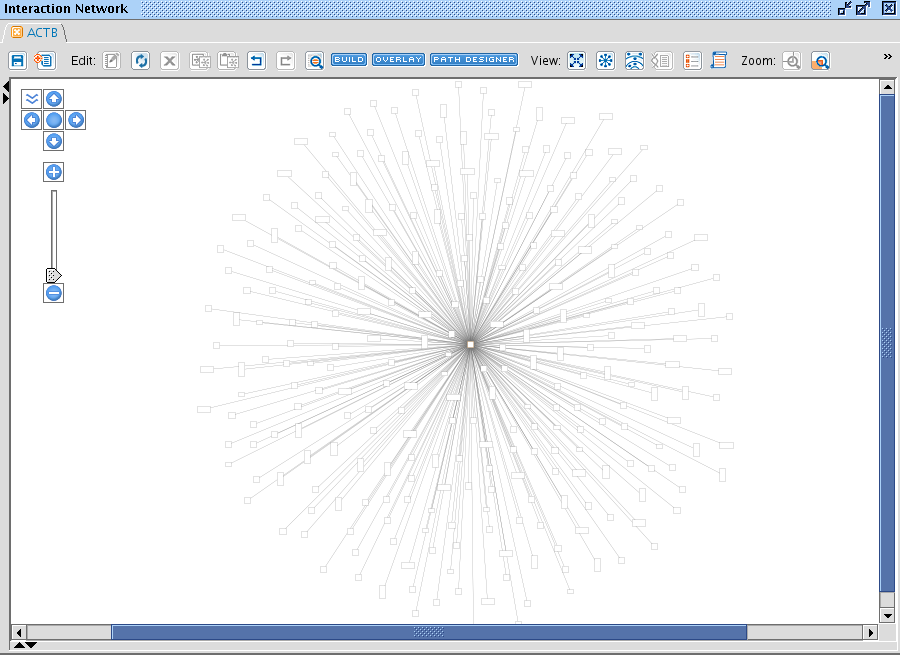
By clicking on > Interaction Network, a new Pathway window opens up and displays your molecule in the middle and all molecules that are known to interact with it.
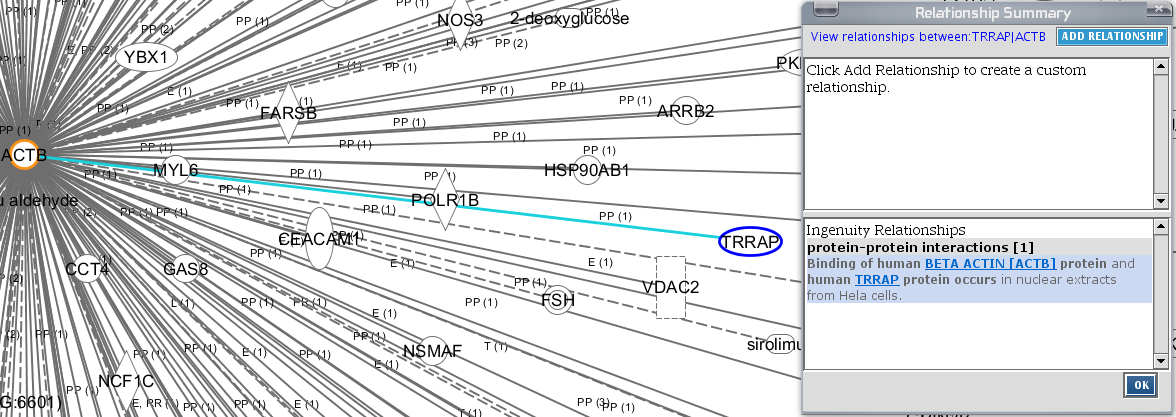
This hairy ball summarizes an awful lot of information. You can browse around (scrolling with the mouse wheel etc) and double-click on any entity or relationship you encounter to obtain more information displayed in a small separated window.