Install Picard tools in Linux
Go back to parent page Introduction to Linux for bioinformatics
Contents
Picard tools
Picard comprises Java-based command-line utilities that manipulate SAM files, used for all kinds of HTS sequencing studies. Picard is one .jar file, which can be executed by JAVA. We will go through the steps to install and run Picard tools.
Download
Download the latest version of Picard from http://broadinstitute.github.io/picard/.
While downloading picard-tools-2.2.1.zip, go back to http://broadinstitute.github.io/picard/ to read the 'Quick Start' .
Unpack the archive
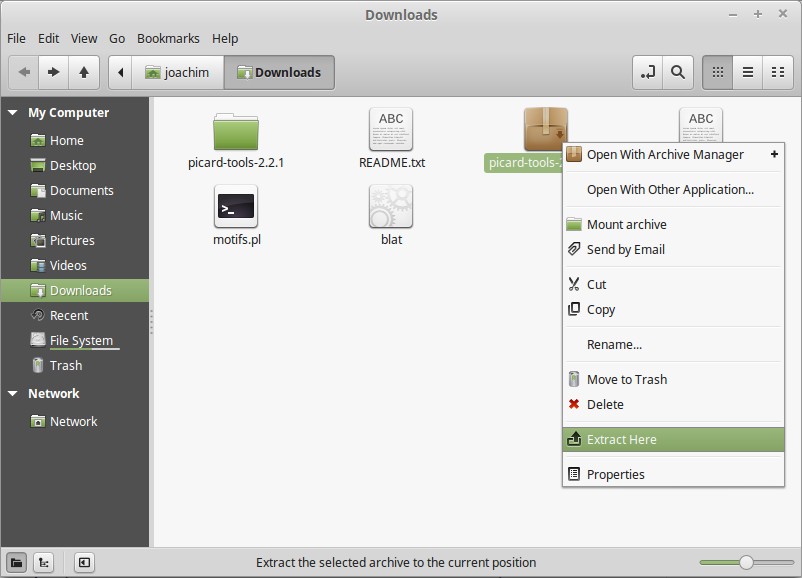
In the ~/Downloads folder, unpack the .zip file.
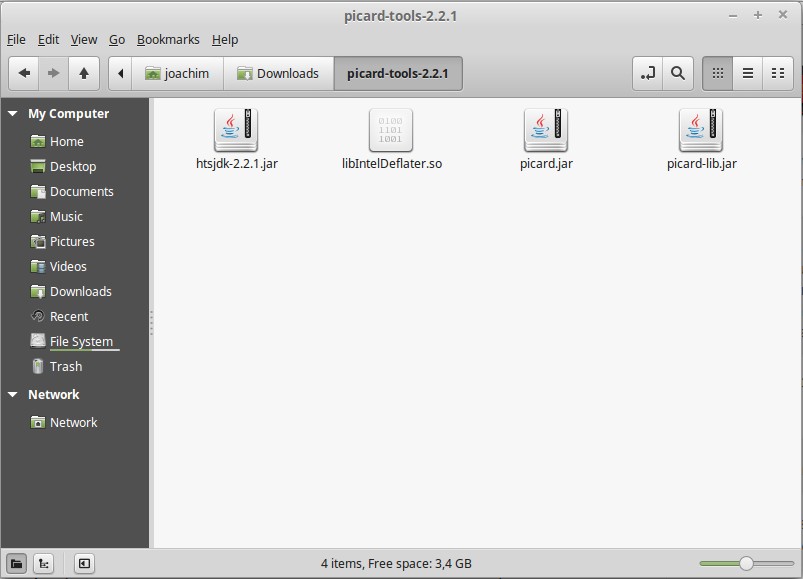
In the unpacked folder, you will find picard.jar.
Check whether the right version of JAVA is present
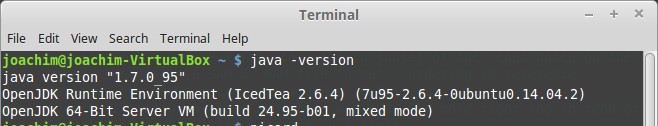
$ java -version
For the latest Picard tools, you need JAVA version 1.8.x (where x is any number). You can try to run with another version, but you will get errors.
Install JAVA 1.8 if necessary
In case you have the wrong JAVA version, you need to install the correct one. Installing JAVA manually is NOT advised.
Googling for 'linux install java 8' gives some leads. For example, this PPA. Install now JAVA 8 from this PPA. The instructions are under the link.
Tip Webupd8.org is a trusted source.
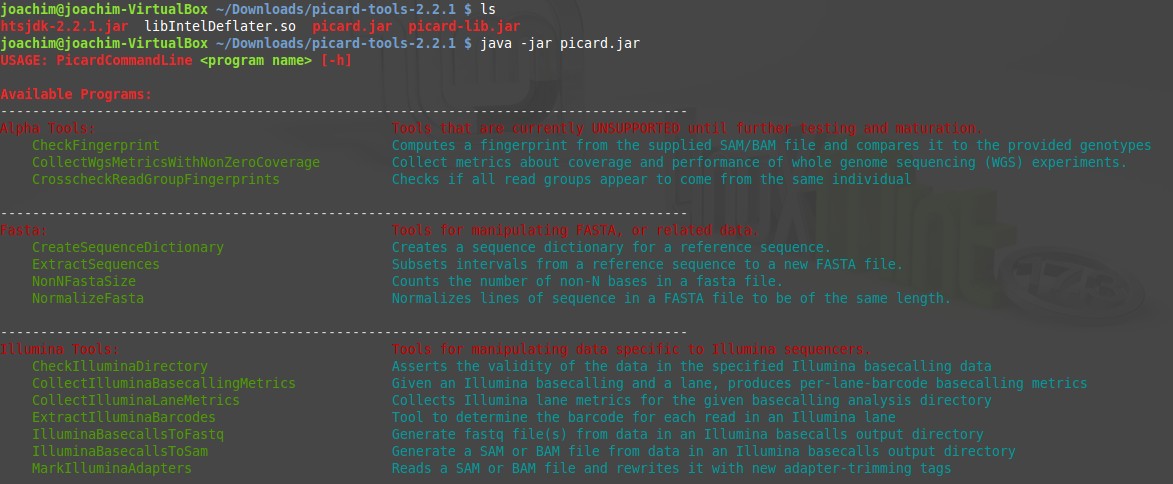
Test run the picard tools
Open the terminal in de Picard-tools directory.
$ java -jar picard.jar
Looks good! Congratulations.
Advanced: efficiently using the Picard tools
Only if you have time left!
To efficiently use this tool in your analyses,
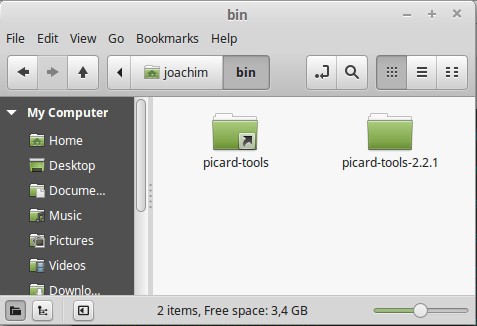
- move picard-tools-2.2.1/ to your ~/bin folder.
- make a symbolic link in ~/bin to the picard-tools-2.2.1 and name it picard-tools
- create an environmental variable 'PICARD' that stores '~/bin/picard-tools/picard.jar (see later in the training)
$ PICARD='/home/joachim/bin/picard-tools/picard.jar'
- make aliases in the file .bashrc (see later in the training)
$ alias picard="java -jar $PICARD"
- Ready to run!
Go back to parent page Introduction to Linux for bioinformatics